Lipids and Lipidomics
Lipids are essential molecules that play crucial roles in energy storage, cell membrane integrity, and signaling processes within biological systems. The study of these vital substances falls under the domain of lipidomics, a field that explores the complex lipid profiles within cells and tissues to unveil their roles in health and disease. NExTLi, the National Expert center for Translational Lipidomics, is dedicated to pioneering research in lipidomics by offering comprehensive services and expertise to researchers aiming to unravel the mysteries of lipid metabolism and its implications.
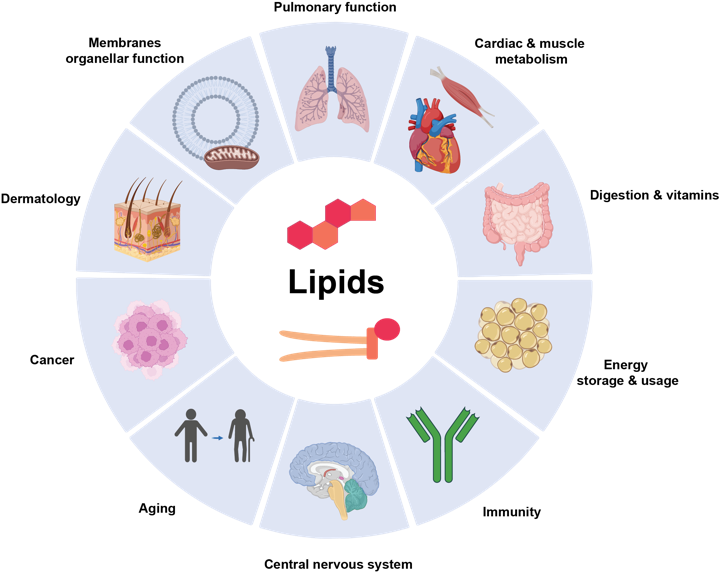
Building upon the foundation of traditional lipidomics in the Amsterdam UMC Core Facility Metabolomics, NExTLi focuses on advancing lipidomics research by exploring the complexity of lipid profiles in health and disease. Our current capabilities enable us to offer a wide range of lipidomics services, including 2D-, 3D-, and 4D-lipidomics. These technologies provide detailed insights into lipid composition, structure, and metabolism, supporting the identification of novel biomarkers and therapeutic targets.
At NExTLi, we are committed to supporting the research community through our specialized lipidomics services. By leveraging our expertise and state-of-the-art technologies, including advanced tools like AcquireX, we offer researchers the resources and knowledge necessary to deepen their understanding of lipid functions and their impact on human health.
Lipidomics
NExTLi provides different lipidomics services that encompass 2D, 3D and 4D lipidomics. We also can do tracer-based lipidomics. But what does this mean? This is explained below.
[Created with BioRender.com]
2D, 3D and 4D lipidomics?
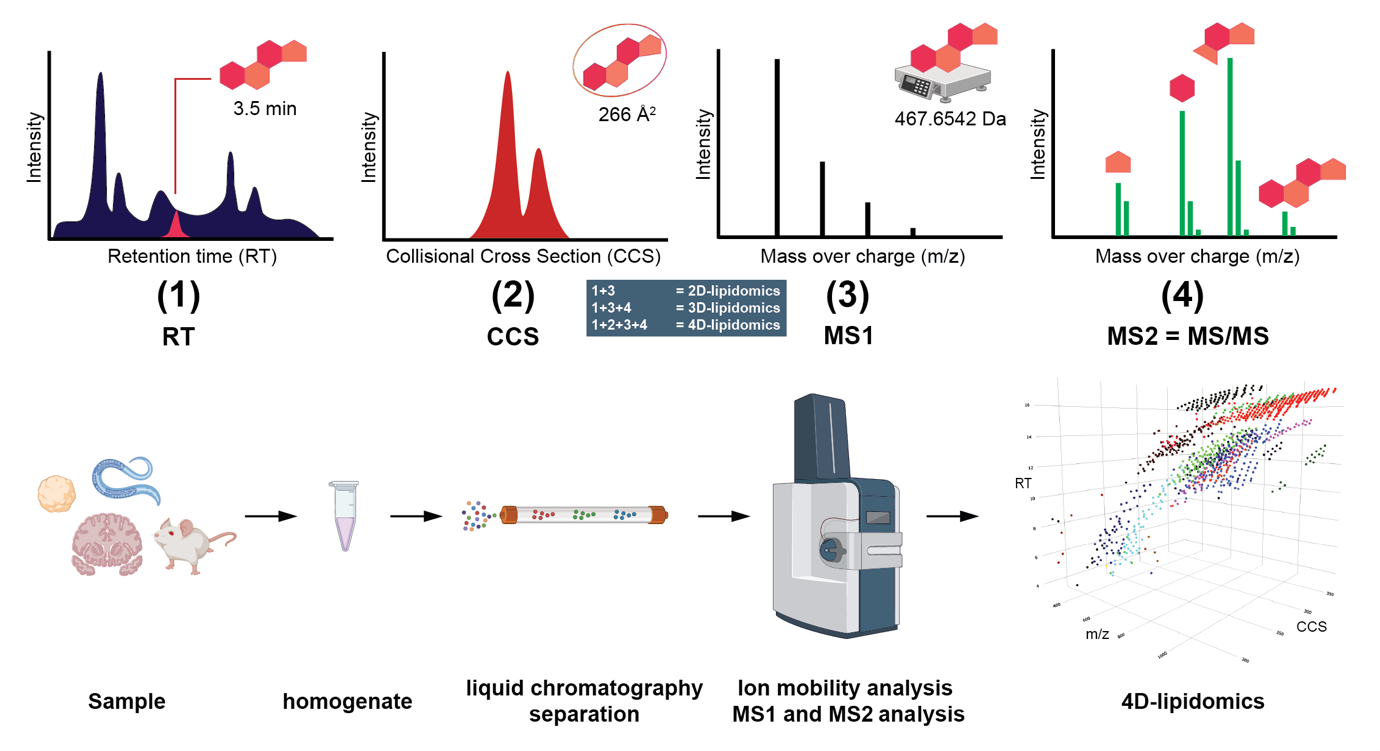
Lipidomics can be performed at different dimensional depths ranging from 2D- to 4D-lipidomics, see Figure 1, where every stage is indicated by a number between brackets. 2D-lipidomics = (1) + (3) uses liquid chromatographic separation of the lipid, yielding retention time = RT = (1), followed by mass spectrometric detection of the lipid (3), termed MS1, expressed as m/z = mass over charge. 3D-lipidomics = (1) + (3) + (4) usually adds fragmentation of the lipid (4), termed MS2 or MS/MS, to the chromatographic and MS1 dimension to gather more structural information, thereby enhancing identification. In 4D-lipidomics (1) + (2) + (3) + (4), ion mobility (2), expressed as the collisional cross section (CCS), is added, which is a physical constant representing the shape/surface area of the molecule. Ion mobility can be used to separate molecules even if they have the same m/z in the MS1 stage. This allows the identification of more lipids at a higher structural level and certainty.
Ion mobility?
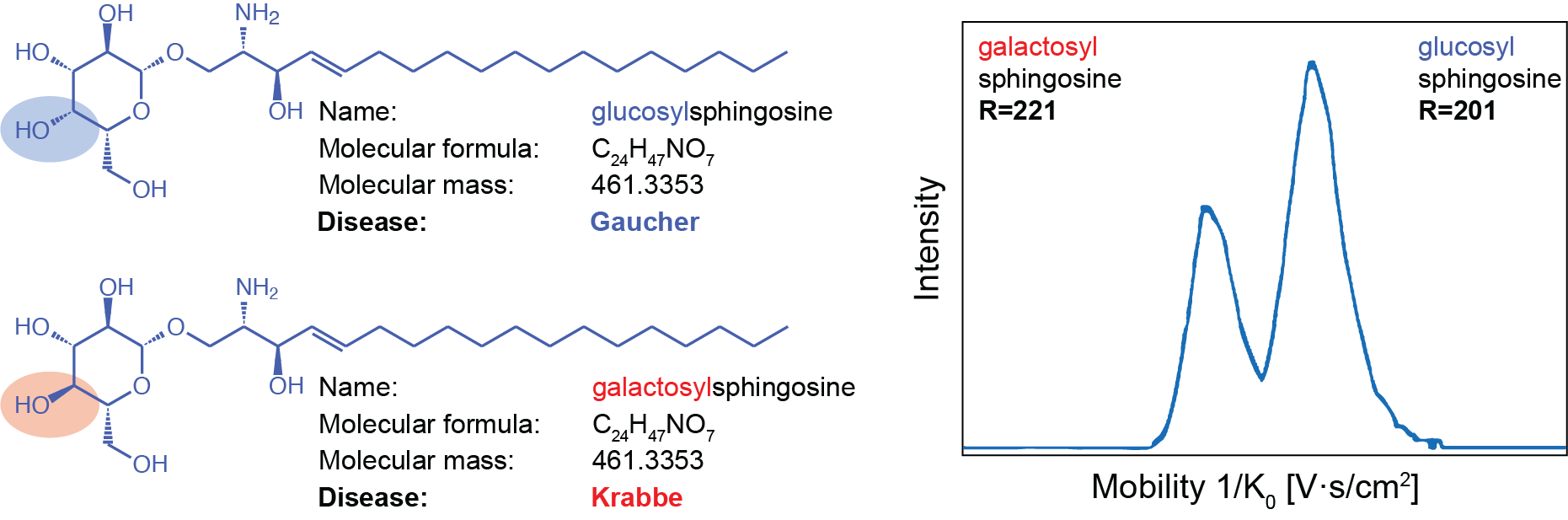
We mention the term ion mobility as one of the dimensions that is used to make the mixture of lipids less complex. Ion mobility is technique that enables the separation of metabolites with the same exact mass but different 3-dimensional conformations. This advanced ion mobility approach is ideally suited for lipidomics as it increases the number of identifiable lipids, enhances compound identification confidence, and in some cases eliminates the need for chromatographic separation, which is especially important for applications where chromatographic separation is not possible, such as mass spectrometry imaging. A nice use-case is presented Figure 2.
Tracer-based lipidomics?
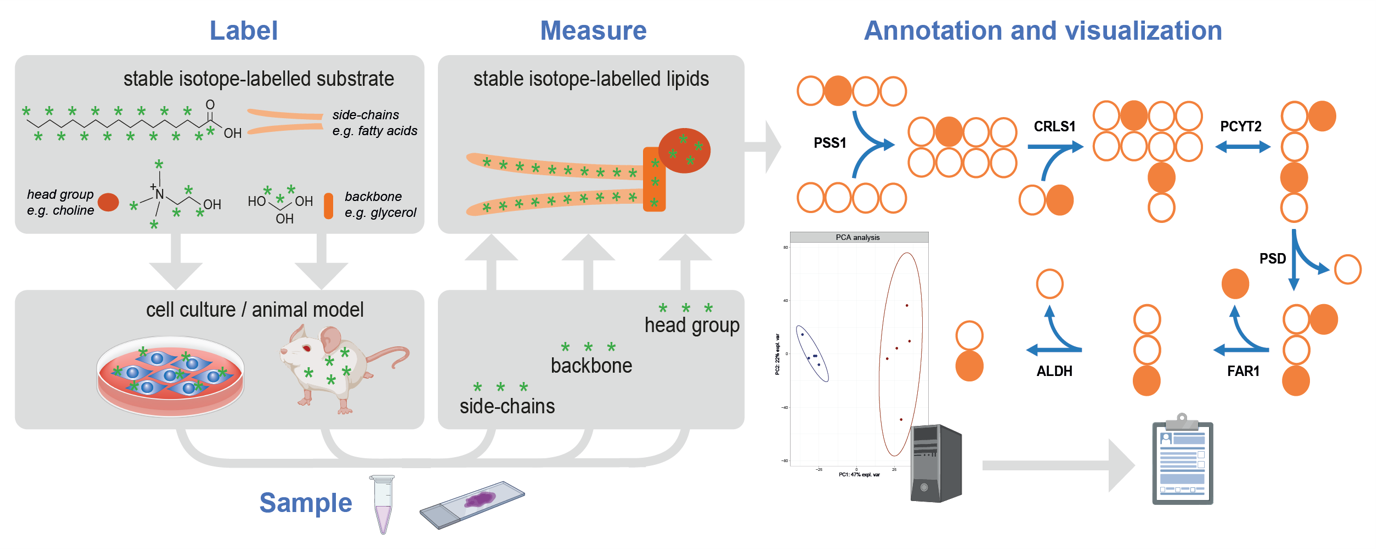
Tracer-based lipidomics and fluxomics utilize non-radioactive stable isotopes to investigate lipid metabolic processes, focusing on the synthesis, modification, and turnover of lipids in biological systems, see Figure 3. These stable isotopes, distinguishable by their heavier mass via mass spectrometry, enable the tracking of lipid molecule changes. Fluxomics builds upon this and tracks changes over time which allows the determination of the rates of metabolic reactions and pathways at the cellular, tissue, or organism level, offering an extensive understanding of metabolic flux and its regulation.
[Created with BioRender.com]
AcquireX?
To further enhance the certainty of lipid annotation, we utilize AcquireX, an innovative platform that automates the selection and fragmentation of lipids during mass spectrometry analysis. By prioritizing the most informative ions for fragmentation, AcquireX reduces redundancy and ensures comprehensive coverage of the lipidome. This targeted approach significantly improves structural identification, enabling us to distinguish closely related lipid species and refine lipid annotations with greater confidence.
Technologies
NExTLi’s technological arsenal is equipped with state-of-the-art instruments and corresponding bioinformatics workflows designed to meet the broad spectrum of research needs within the field of lipidomics. Our facility harnesses cutting-edge analytical tools to provide comprehensive lipid analysis, detailed lipid profiling, and pioneering spatial lipidomics services.
Instrumentation
Mass spectrometers for lipidomics: Our core capabilities include four high-resolution Orbitrap mass spectrometers and the innovative timsTOF Pro 2 system featuring advanced ion mobility. These platforms facilitate 2D, 3D, and 4D lipidomics analyses, offering unparalleled sensitivity, specificity, and throughput for the quantitative and qualitative analysis of lipidomes. In addition, we have 8 triple quad mass spectrometers and 2 GC(MS)s that also can be used for (targeted) lipid measurements, such as total fatty acids.

Advanced bioinformatics and data processing
NExTLi stands at the forefront of bioinformatics and data processing within the lipidomics field, integrating sophisticated computational strategies to decipher complex lipid data. Our team harnesses custom-developed algorithms and state-of-the-art software to analyze, interpret, and visualize lipidomics datasets, enabling precise identification and quantification of lipid species across various biological samples. With a focus on enhancing the accuracy and efficiency of lipidomics research, we provide researchers with comprehensive data analysis support, from experimental design to insightful biological interpretation. Our bioinformatics infrastructure is designed to manage large-scale lipidomics data, applying advanced statistical methods and machine learning techniques to uncover novel lipid biomarkers and metabolic pathways. At NExTLi, we are dedicated to pushing the boundaries of what’s possible in lipidomics data analysis, offering researchers powerful tools to drive forward their scientific inquiries.
Through the synergistic use of these instruments and bioinformatics/data processing workflows, NExTLi is at the forefront of lipidomics research, offering unparalleled expertise and services to the scientific community. Our commitment to technological advancement and precision analysis positions us as a leader in the exploration of lipid function and metabolism, propelling the field toward new scientific discoveries and therapeutic approaches.
